Integrating Electroanatomic Mapping Data with EP Data
Table of contents
- Background
- Quickstart
- Notes on exporting data
- Functions
- Return all the Carto points which have Bard data associated with them
- Get a single Bard electrogram associated with a Carto point
- Get a single filtered Bard electrogram associated with a Carto point
- Change the filter settings for the electrograms
- Plot the electrogram, filtered electrogram and location for one of these points
- Plot a map showing the positions of all the bard electrograms
- Get all the valid Carto points associated with a particular Bard file
- Get all the Bard electrograms, and their associated positions, dependent on whether or not a channel was being paced
- Troubleshooting
- Publications
Background
A common use-case is to record electrograms for an extended period using an electrophysiology recording system such as the Bard, whilst documenting anatomical recording sites using electroanatomic mapping platform such as Carto.
This section documents a class, Userdata, which can be used to perform this data integration, and exposes helpful functions for subsequent data processing.
Quickstart
barddir = '/Volumes/Extreme SSD-confidential/CETT-AF Dominant Frequency Study/Data/Case 1';
load('/Volumes/Extreme SSD-confidential/CETT-AF Dominant Frequency Study/Data/Case 1/Study_1331_04_26_2020_17-59-21_1-LA-DF_Points.mat')
u = Userdata(userdata, barddir);
Notes on exporting data
The Userdata class accepts two inputs:
- A
userdatastructure, such as created by cartodatareader. barddir, an absolute path to a directory of bardfiles
By necessity, a lookup table linking the bard files to the Carto points is necessary. To avoid additional files; this lookup table is provided in the bardfile file names, using the following conventions:
BardFile, ChPace'Map d', P1820-1829, [Pacing LAA].txt
- All files should start ‘BardFile …’
- Filename sections are separate with commans, by default, although technically this does not matter
- All files should be stored in the same directory
- Relevant notes; for example catheter moved, etc; should be added between square brackets. Square brackest must not otherwise be used in the filename
- The related Carto points must be specified as Pi-j, where i is the index of the first point and j the index of the second point
- The paced channel should be specified between single quotes (‘) following the text ChPace
Functions
Return all the Carto points which have Bard data associated with them
points = u.listPointsWithBardElectrograms();
Get a single Bard electrogram associated with a Carto point
egm = u.getBardEgm(points(1));`
Get a single filtered Bard electrogram associated with a Carto point
egm = u.getBardFiltEgm(points(1));`
Change the filter settings for the electrograms
set(u.bardFiles, 'Filter', [100 450]);`
Plot the electrogram, filtered electrogram and location for one of these points
figure
subplot(2,2,[2 4]); u.drawBardElectrogramSites(points(1))
subplot(2,2,1); plot(u.getBardEgm(points(1))); title('elecgrogram');
subplot(2,2,3);
plot(u.getBardFiltEgm(points(1)));
title('filtered elecgrogram');
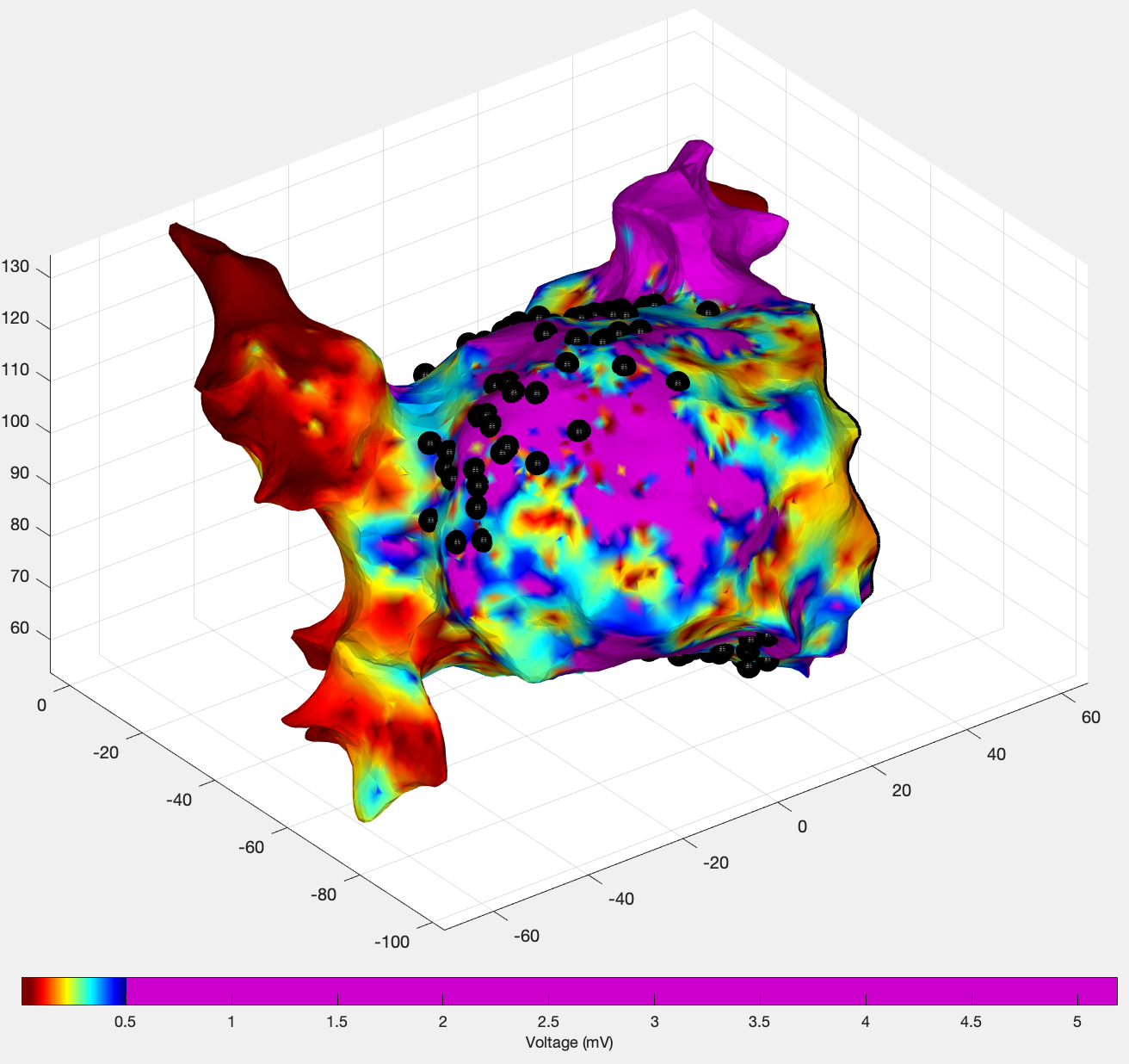
Plot a map showing the positions of all the bard electrograms
u.drawBardElectrogramSites(':');
Get all the valid Carto points associated with a particular Bard file
u.getValidCartoPointsAssociatedWithBardFile(1)
Get all the Bard electrograms, and their associated positions, dependent on whether or not a channel was being paced
tbc
Troubleshooting
This class requires that BardFile accepts PARAMETER-VALUE pairs. This functionality is available on branch feature/command_line_input_only (April 2020)
Issue 1
Publications
This code has been used in the following publications.